November 2018—CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Weill Cornell Medicine. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Jordan Baum, MD
Rebecca Marrero, MD
Alain Borczuk, MD
Hanna Rennert, PhD
Next-generation sequencing is becoming the standard of care in the diagnostic workup of lung adenocarcinoma and other solid tumors. This technology leverages massively parallel sequencing to interrogate multiple genes of interest in a single test. The results of NGS have important implications for patient care, providing diagnostic, prognostic, and predictive information.
False-positive NGS results may arise due to multiple scenarios, for example, misidentification of a germline finding as a somatic finding. These false-positive results may lead to misinterpretation or inappropriate use of the NGS results and have serious clinical impact. Here, we discuss a case of NGS solid tumor testing revealing mutations from the patient’s concurrent hematologic malignancy.

Case. An 82-year-old female with chronic myelomonocytic leukemia (CMML) and a remote history of tobacco smoking presented to the emergency department with dyspnea and cough for several weeks that had failed to improve with antibiotics. A chest x-ray and subsequent CT scan without contrast were performed, revealing a 2-cm right upper lobe spiculated lesion. Lung adenocarcinoma was diagnosed on fine-needle aspiration of the nodule, leading to a lobectomy resection of the patient’s right upper lobe. Histologic evaluation of the resection specimen revealed a poorly differentiated pleomorphic carcinoma consisting of adenocarcinoma and giant cell carcinoma with visceral pleura involvement and lymphatic invasion (Fig. 1).
Targeted next-generation sequencing was performed following microdissection of formalin-fixed, paraffin-embedded tissue from the FNA cell block and resection specimens using the ion semiconductor-based sequencing platform Ion Torrent Personal Genome Machine (Thermo Fisher Scientific) with the Ion AmpliSeq Cancer Hotspot Panel v2 (Thermo Fisher Scientific). The panel concurrently interrogates 2,800 hotspots/variants with 207 amplicons in 50 cancer-related genes. Sequence data analysis and variant calling were performed with Torrent Suite Software 5.0 (Thermo Fisher Scientific).
The cytology specimen had a visually estimated tumor cellularity (or neoplastic content) of 25 percent, while the surgical pathology resection specimen had a higher tumor cellularity of 70 percent. Sequencing of the lung cancer specimens revealed multiple concurrent mutations as follows (Table 1):
- c.34G>T missense mutation (NM_004985, p.Gly12Cys) in the KRAS gene with 22 percent variant allele frequency (VAF) in the FNA and 38 percent in the resection;
- c.35G>A missense mutation (NM_004985, p.Gly12Asp) in the KRAS gene with three percent VAF in the FNA and six percent in the resection;
- c.35G>A missense mutation (NM_002524, p.Gly12Asp) in the NRAS gene with 15 percent VAF in the FNA and three percent in the resection; and
- c.52delA frameshift deletion (NM_000546, p.Thr18Hisfs*26) in the TP53 gene with 23 percent VAF in the FNA and 41 percent in the resection.
Due to the finding of multiple concurrent mutations in cancer driver genes (KRAS and NRAS) on NGS testing of the lung adenocarcinoma, the patient’s clinical history regarding her CMML was investigated further. The patient had a reported history of long-standing mild monocytosis since 2009 and elevated hemoglobin and hematocrit since 2013, which were being followed by a hematologist. However, the patient refused to undergo a bone marrow aspiration. In 2015, when the patient was 80 years old, a follow-up complete blood count revealed leukocytosis (14.2 × 103/µL [ref: 3.4–11.2 × 103]) with 15.4 percent monocytes (ref: 2.0–11.0 percent). The patient then agreed to a bone marrow aspiration, which revealed monocytosis without an increase in blasts or dysplasia. Cytogenetics showed a normal female karyotype. A limited, targeted, amplicon-based NGS panel for myeloproliferative neoplasms interrogating five genes was performed at Genoptix (Carlsbad, Calif.) on a peripheral blood sample, which was negative for mutations in JAK2, MPL, CALR, CSF3R, and SETBP1. Because cytogenetics and the limited MPN panel failed to reveal an acquired clonal cytogenetic or molecular genetic abnormality, a full myeloid molecular profile using amplicon-based NGS technology interrogating clinically relevant regions of 44 genes was additionally performed (Genoptix) and revealed the following (Table 1):
- c.2278C>T nonsense mutation (NM_015338, p.Gln760*) in the ASXL1 gene with a VAF of 43 percent; and
- c.35G>A missense mutation (NM_002524, p.Gly12Asp) in the NRAS gene with a VAF of 34 percent.
The patient was then followed clinically for her CMML and did not receive any therapy or further treatment. She had an absolute monocytosis of 1.6 × 103/µL (ref: 0.2–0.9 × 103/µL) one week prior to the FNA and 2.6 × 103/µL the day of the right upper lobectomy.
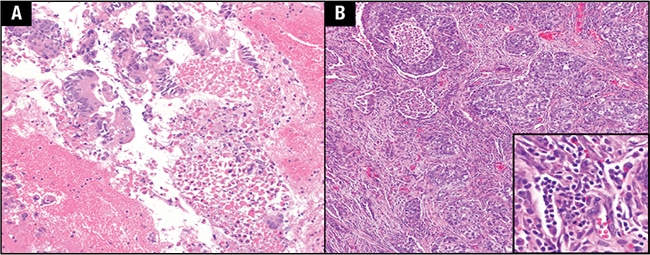
Fig. 1. A. Fine-needle aspiration cell block of the lung nodule. B. Low-power view of the lung nodule resection specimen. Inset shows numerous leukocytes packing intratumoral blood vessels on high-power view.
We speculated that additional mutations may have been present on the peripheral blood sample for the CMML workup but not reported in the official Genoptix report. The Genoptix pathologist responsible for interpreting the patient’s case was contacted to determine if variants were identified in the sample that were not reported. It was verbally confirmed that an additional mutation was present, a c.35G>A missense mutation (NM_004985, p.Gly12Asp) in the KRAS gene with a VAF of three percent (below their laboratory minimum quality control metric of VAF for reporting, five percent). The TP53 gene is included on the Genoptix myeloid molecular profile panel; however, no TP53 variants were identified in the patient’s sample. Based on the findings in the patient’s next-generation sequencing results from the lung adenocarcinoma, the patient was not eligible for targeted tyrosine kinase inhibitor therapy and received standard chemotherapy with carboplatin and pemetrexed.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
